By Tyler Benster
Neuroscientists have a dizzying array of methods to listen in on hundreds or even thousands of neurons in the brain and have even developed tools to manipulate the activity of individual cells. Will this unprecedented access to the brain allow us to finally crack the mystery of how it works? In 2017, Jonas and Kording published a controversial research article, “Could a Neuroscientist Understand a Microprocessor?” that argues maybe not. To make their point, the authors turn to their “model organism” of choice: a MOS 6502 processor as popularized by the Apple I, Commodore 64, and Atari Video Game System. Jonas and Kording argue that for an electrical engineer, a satisfying description of the processor would break it into modules, like an adder or subtractor, and submodules, like the transistor, to form a hierarchy of information processing. They suggest that, while popular methods from neuroscience might reveal interesting structure in the activity of the brain, researchers often use techniques that would fail to reveal a hierarchy of information processing if applied to the (presumably much simpler) computer processor.
For example, neuroscientists have long used lesions, or turning off or destroying a part of the brain, to try to find links between that brain region and particular behaviors. In one particularly striking experiment, the authors mimicked this classic technique by simulating the processor as it performed one of four “behaviors”: Donkey Kong, Space Invaders, Pitfall, and Asteroids. They then systematically removed one transistor, and reported which (if any) of the behaviors could still be performed (i.e. did the game boot?) The elimination of 1,565 transistors have no impact, while 1,560 inhibit all behaviors, and indeed a subset of transistors make only one game impossible. Perhaps these are the Donkey Kong transistors, the authors coyly suggest, before concluding that the “causal relationship” is highly superficial. Given that the design of the microprocessor does not designate certain transistors for use in particular programs, is it reasonable to conclude that a particular transistor would only be involved in running Donkey Kong?
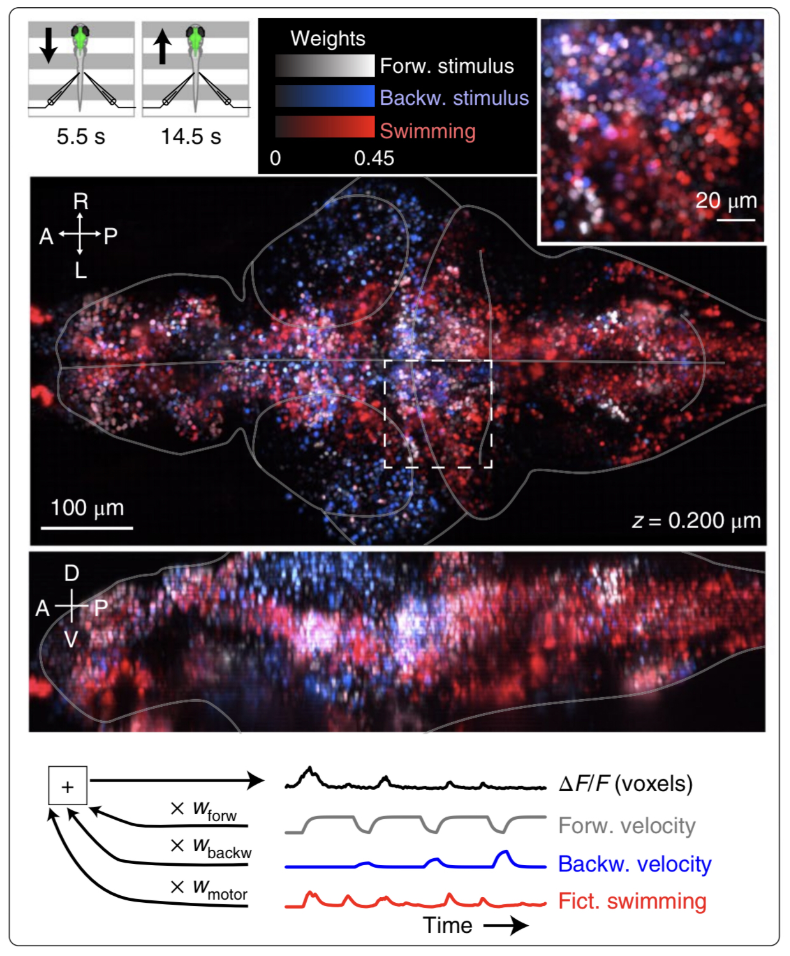
Figure 1: Neuroscientist Nikita Vladimirov and colleagues combine a sophisticated technique with a clever behavioral task to investigate the larval zebrafish brain. By moving spatial gratings forwards or backwards under the animal, the scientists simulated the feeling of moving water, driving the zebrafish to try to swim to stay in place. While the animal was swimming, the scientists watched as thousands of neurons in the brain changed their activity. They could then relate particular activity patterns to particular behaviors—for example, here all the neurons related to forward velocity are colored white, backward velocity are blue, and swimming are red. In this way, the scientists could see how different parts of the brain might contribute to particular behaviors.
This brings us to a new paper by Vladimirov, et al, wherein the authors use a powerful imaging method, light sheet microscopy, to observe the activity of nearly all the neurons in a larval zebrafish while the animal performs a simple task (figure 1). The task leverages the larval zebrafish’s natural instinct to maintain its position in a moving stream, which the authors simulate by moving a spatial grating backwards or forwards beneath the animal. The animal infers from the moving gratings that the water is moving and tries to remain in the same location, responding with vigorous tail movements as the grating moves forwards and reducing tail movements as the pattern moves backwards. By imaging the brain during this innate behavior, the authors are able to “map” neurons that are highly correlated with forward stimulus, backward stimulus, or tail movements. The authors then chose a handful of small regions with similarly tuned neurons, and lesioned these cells using short, spatially-targeted pulses of a laser. Among groups of motor-tuned neurons, this resulted in highly variable changes in behavior. Some of these changes were expected, such as a decrease in motor vigor after ablation of neurons involved in arousal control and generation of swim bouts. Others were more surprising: swim frequency actually increased after ablation of an area thought to be important for swimming and posture control.
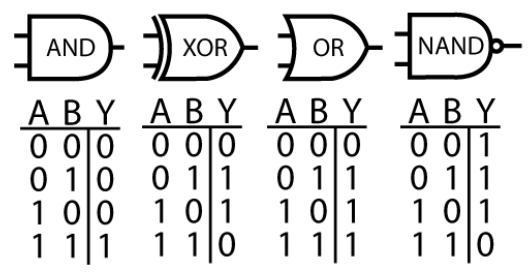
Figure 2: Computer processors can be understood in terms of logic gates; i.e. what operation each small group of transistors performs. Is it possible to conceptualize neurons of the brain in the same way?
This result illustrates a surprising difference between the brain and computers: whereas in a computer, correlation between the activity of a transistor and a particular program is likely to predict the transistor’s necessity, the same is not true for the brain. Neurons correlated to a task may or may not be necessary, and are more likely to subtly modify behavior rather than the coarse crash-or-run dichotomy of computers. As such, perhaps we learn more by mapping modules in brains versus computers. Nonetheless, it is difficult to see how the lesion experiment passes the sieve proposed by Jonas & Kording: would performing this experiment on a microprocessor tell us anything about how it functions? The answer, as performed in a circuit-wide (read: brain-wide) fashion, is no. However, Vladimir et al. go on to perform a series of experiments not considered by Jonas & Kording: the direct stimulation of small groups of functionally-defined neurons, producing activation maps that suggest causal connectivity of neurons.
Imagine if such a technique were applied to a single transistor: manipulating the voltage of the transistor source and gain while monitoring the drain. We would quickly learn how a transistor works. Next, we might try observing and perturbing a handful of transistors, and learn the logic gate primitives: AND, XOR, OR and NAND (figure 2). Then, we might scale to a small circuit, like an adder or an accumulator. If we could impose state on the circuit and observe its output, we would be able to deduce the function of the module. By systematically performing this task on all modules, we could functionally describe the pieces of the whole. Finally, with the connectome of the circuit, we might arrive at a hierarchical description that would satisfy electrical engineers. If we try to apply this approach to the brain starting with a single neuron, arguably the closest equivalent to a transistor, we might feel reasonably satisfied with our biophysical understanding of a neuron thanks to the work of Hodgkin-Huxley. But scaling to the next level of abstraction brings a much tougher question: what are the modules of the brain? We do not know if the brain, like the microprocessor, can be summarized by a hierarchical structure.
References:
-
Jonas, Eric & Kording, Konrad Paul. “Could a neuroscientist understand a microprocessor?” PLOS CompBio, 13(1): e1005268 (2017).
-
Vladimirov, Nikita et al. “Brain-wide circuit interrogation at the cellular level guided by online analysis of neuronal function.” Nature Methods, 15: 1117-1125 (2018).