Q&A: New imaging tool unravels the brain's complex machinery in health and disease
In Alzheimer’s disease, abnormal proteins snake through the brain, causing neurons to wither away and die. But even as these proteins choke out the brain’s molecular machinery, some Alzheimer’s patients live well into old age without showing any symptoms. Understanding what makes some brains resilient in the face of neurological diseases like Alzheimer’s could be a key to future treatments.
But decades of research have failed to extract clear answers from the brain’s fiendish complexity. Neural tissues are built from the interactions between a hundred or more specialized kinds of cells, each of which are powered by thousands of intricate molecular machines. Understanding these complex systems — and where they go wrong in neurodegenerative disease — requires a way of visualizing these interlocking components of brain tissue, all at the same time.
Wu Tsai Neurosciences Institute affiliate Sean Bendall thinks he has just the tool for the job. A firm believer that many stubborn scientific questions just require a new way of looking at the problem, Bendall heads a team of “unapologetic tool builders” constructing powerful cell and tissue imaging technologies to help untangle biology’s molecular complexity. The team’s latest technique — called multiplex ion beam imaging, or MIBI — provides a new way to simultaneously visualize dozens of different brain tissue components in potentially millions of combinations.

Sean Bendall. Associate Professor in the Department of Pathology
Devised in the lab of immunologist Garry Nolan, where Bendall trained before taking charge of his own team, MIBI uses an ion beam to scan back and forth across a tissue specimen, releasing metal-tagged antibodies that mark the location of dozens of molecules of interest. Building on an earlier single-cell multiplexing technique Bendall helped pioneer in the Nolan lab, called cytometry by time of flight (CyTOF), MIBI uses mass spectrometry to identify these metal ions in flight, building a high-dimensional molecular portrait of a piece of intact tissue with nanometer-scale resolution.
MIBI complements other multiplexed tissue imaging techniques available to Stanford researchers, such as CODEX — another technique developed by the Nolan lab that tags tissue samples with fluorescent markers before reading out their molecular characteristics. With these other techniques, imaging the proper fluorescent markers can sometimes be a tricky task due to the natural fluorescence of tissue, which can impede other fluorescent signals. In contrast, MIBI’s use of mass spectrometry instead of fluorescent markers allows it to avoid some of these limitations, aiding the Bendall lab’s mission to help researchers study the complexities of the human brain.
Bendall’s team, which is based in the Department of Pathology, has published multiple studies on MIBI’s use in cancer and immunology. But the team’s most recent study, currently available as a preprint on BioRxiv, trains MIBI on one piece of the resilience puzzle in Alzheimer’s.
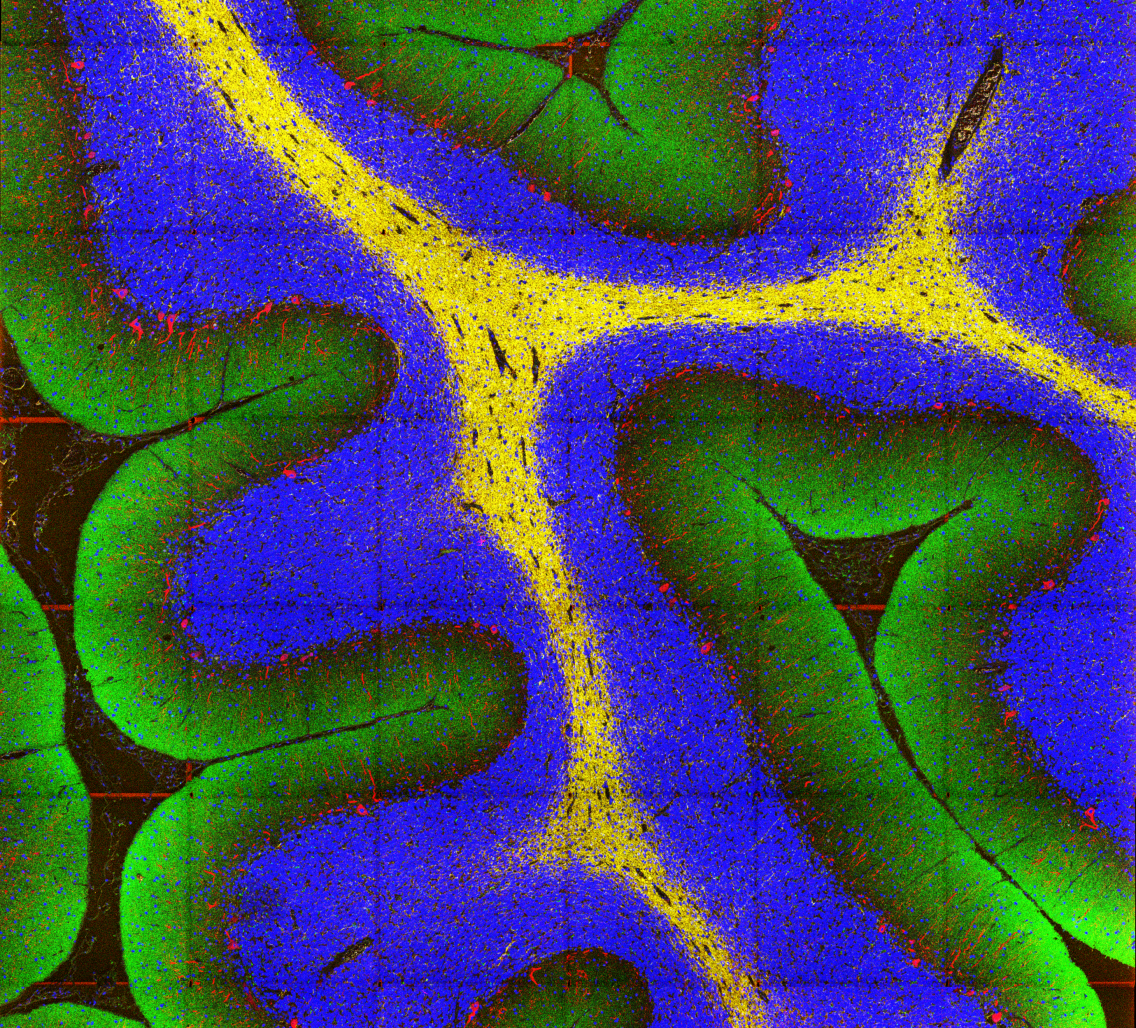
Using MIBI’s multiplexing capabilities, the team explored why the same molecular gunk characteristic of Alzheimer’s might be seen in the brains of both seemingly disease-free and diseased patients. The team looked to define pathological regions in both types of brains, seeking patterns across preserved postmortem brain samples from patients at different cognitive levels. One finding in all patients: small groups of persistently healthy neurons characterized by increased expression of the protein MFN2, a protective factor that’s been linked to Alzheimer’s disease in the past, but has resisted comprehensive analysis.
The study, Bendall said, is a proof-of-concept for applying the power of multiplexed imaging in neuropathology. Bendall envisions MIBI as a tool to be trained on key questions facing not only Alzheimer’s research, but neurodegenerative disease as a whole. Bendall has worked to make MIBI more accessible to the scientific community, founding Ionpath, a company that aims to make MIBI commercially available to clinical researchers across the world. And at Stanford, the Human Immune Monitoring Center plans to make a new MIBI tool available next year for use by Stanford researchers, funded by the university’s c-ShARP program.
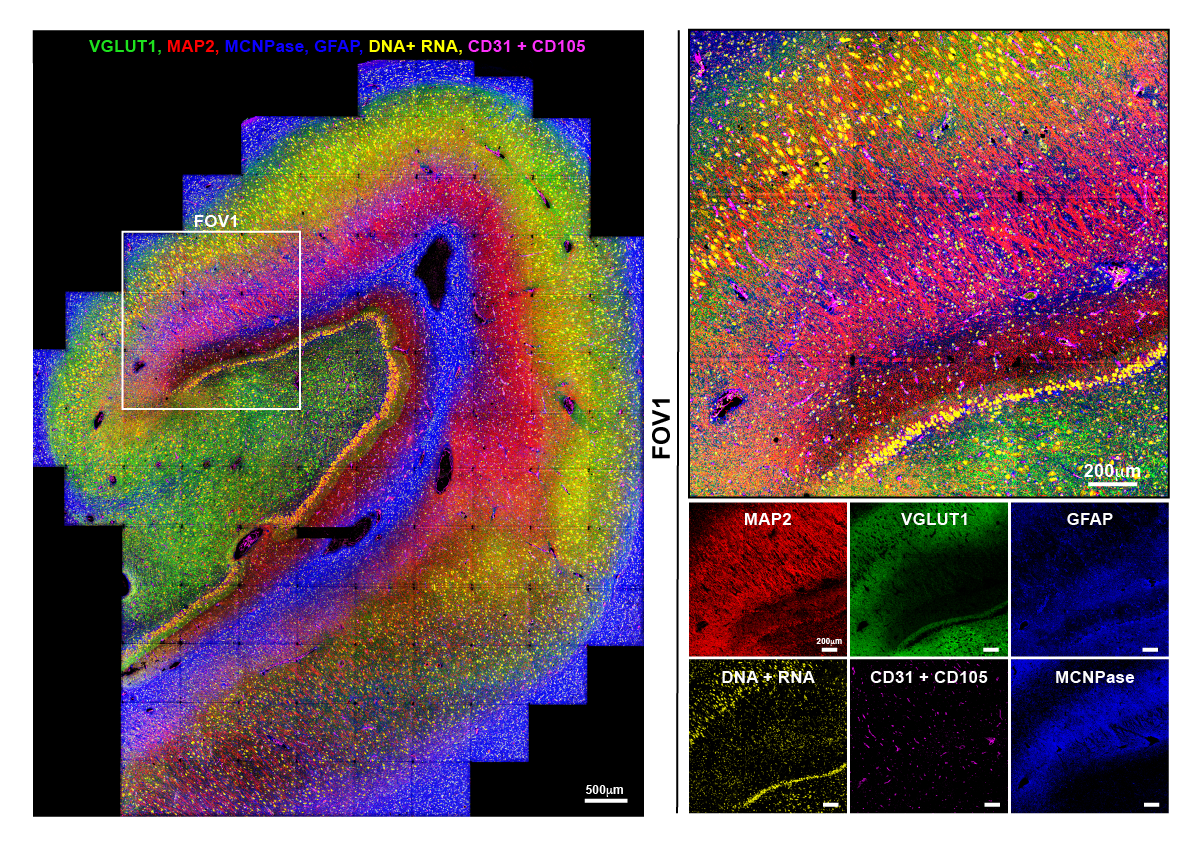
Proof of concept image of human hippocampus tissue showing just eight of the 38 different protein markers researchers were able to image on this piece of tissue. Shown here in five colors are VGLUT1 (areas of synaptic density), MAP2 (neuronal dendrites and soma), MCNPase (myelin), GFAP (astrocytes), DNA & RNA (showing nuclei and neuronal soma) and CD31 & CD105 (vasculature). In the boxed FOV1 region of interest, these markers reveal larger areas of myelin, astrocytes and synaptic density as well as the fine structures of dendrites, soma of hippocampal pyramidal neurons and granule cells of the dentate gyrus, nuclei and vasculature. Images courtesy Dunja Mrdjen
We spoke to Bendall to learn more about MIBI and his team’s hopes for how the tool can advance Stanford neuroscience research.
You’ve recently been working to bring your new MIBI technology to the field of neuropathology. How can MIBI help address big questions in neuroscience that couldn’t be addressed before?
It's really a chicken and the egg problem. It’s hard for people to imagine what questions they can ask, if they don’t know that certain technologies exist to help them ask those questions.
If we go back to human biology, we know more about the composition and roles of cells within the haematopoietic system [the system involved in the creation of blood cells] and the immune system than any other tissue in the body. This is mainly driven by advances in cytometry, which has bolstered our ability to look at these systems as a collection of different pieces. This technology has helped us figure out what those pieces are and analyze them.
Now, compare our elegant understanding of those systems with the brain, where we are much more limited when it comes to measuring neurological regulation and disease. Unlike the immune system, we have to choose what to study when we do an experiment. Am I going to look at neurons? Am I going to look at the blood brain barrier? You’re not afforded the opportunity to look at all the components at the same time and see how they’re interacting together, and that limits our ability to ask the big, important questions.
Could you elaborate on why we need to look at all of these components at the same time? Why is that aspect of MIBI so important?
Scientists are often not exactly right about what they ‘hypothesize’ is driving a given biological or disease process. If you limit your investigation to just the most likely candidate, you’ll often miss the real answer.”
For example, let’s say that I think that some particular cell type may be responsible for this specific disease’s behavior, and all my experiments shine light on just this one cell type. That’s a problem, because I’m often wrong and nothing in a biological system acts in isolation. Anything I discover from the experiment is going to only apply to that specific cell type while I have ignored everything else.
MIBI lets you take a more holistic approach to any disease or tissue by looking at all the major components and processes simultaneously. The underlying goal is to use MIBI to look at a spectrum of neurodegenerative disease, looking at the central nervous system as a whole, and quantify all of its components. In other words, we’re making a comprehensive model of what’s going on.
How does your new preprint demonstrate the potential capability of MIBI in neuroscience research?
This first paper was meant to serve as a framework for how to use MIBI in neuroscience. We’re laying out our manifesto about how we’re going to take the tools available to us in hematology and immunology, which provide a more holistic picture of those systems, and apply it to neurodegenerative disease and neuropathology.
One of the challenges with studying neurological disease in humans is that you’re not getting biopsies of people’s brains while they’re alive, which means that getting highly qualified brain tissue is difficult. Typically, this is extremely technologically limiting. These tissues often come in an archival format, meaning that the brain is literally dumped in a bucket of formalin and embedded in wax. MIBI is specialized for tackling sample formats like this, which is why we were particularly interested in using it to help study neurological disease.
Looking at your publication history, your lab has touched on a lot of different areas of biology, now including neuroscience and neurodegenerative disease. How do all these things hang together? How would you describe your lab’s mission?
We’re unapologetic tool builders. We build a lot of different platforms and assays to look at cells and tissues at the protein level that people haven’t been able to do before. More recently, we’ve started to delve into applying some of the tissue imaging technologies we’ve developed to look at things like neurodegenerative disease.
What appeals to you about making new tools?
So, my favorite area of science (that isn’t the science that I do) is astrophysics. I love this idea that there’s a lot of known unknowns, and that researchers can build a tool to point at those unknowns and ask a new set of questions.
I’ve also always liked technology. And combining that with my increasing background in cell biology and human pathology, I’ve tried to understand what the challenges are in those fields. In human medicine, what are the unknowns? What do I know about technology and how can we come up with a light to shine on those unknowns and answer those questions?
What are your hopes for future use of MIBI at Stanford?
We’re hoping that this will start to grow as a community resource, the same way that CyTOF has. Short term, we would like to garner interest and institutional support so people know this is here. Long term, and especially with this particular paper, we would like to help institute members understand how MIBI could be used to advance their research.