Mapping the fruit fly: researchers publish a complete Drosophila single-cell atlas

With temperatures warming up in the Bay Area, many of us are familiar with the cloud of tiny, grey flies that begin to colonize our trash bins and fruit bowls. Despite their unassuming appearance, these humble, red-eyed fruit flies—known to the scientific community as Drosophila melanogaster—have made outsize contributions to our knowledge of the brain, embryonic development, and genetics for over a century.
Now, with the publication of the first comprehensive single-cell transcriptional atlas of Drosophila on March 4, 2022 in Science — part of a consortium led by Wu Tsai Neuro scientists and colleagues around the world — our understanding of this diminutive scientific heavyweight is reaching new heights.
Fruit flies are behind five Nobel Prizes on topics as varied as how genes are passed on, why X-rays are dangerous to cells, how genes control early embryonic development, and how the innate immune system gets activated. They may not look much like us, but ~63% of the protein-coding genes in the Drosophila genome have human counterparts. This high degree of functional concordance means that the breakthroughs we make in, for example, understanding how the fruit fly brain is put together can further our understanding of how human brains work. Drosophila is even being used as a model for neurological disorders and mental illness, among other diseases.
Given their widespread use as a model organism, hundreds of publicly available datasets are available profiling the genome, transcriptome, and proteome of this small fly. In particular, single-cell sequencing technologies have been applied to many different Drosophila tissues and developmental stages to produce a wealth of single-cell-resolved transcriptomic data. However, the heterogeneity of all of these datasets—flies of different genetic backgrounds, sequenced using different protocols and platforms, and analyzed by different labs—makes it difficult to directly compare gene expression.
To address this issue, the international Fly Cell Atlas Consortium was formed with the goal of making a complete cell atlas of the entire adult Drosophila on a single sequencing platform.
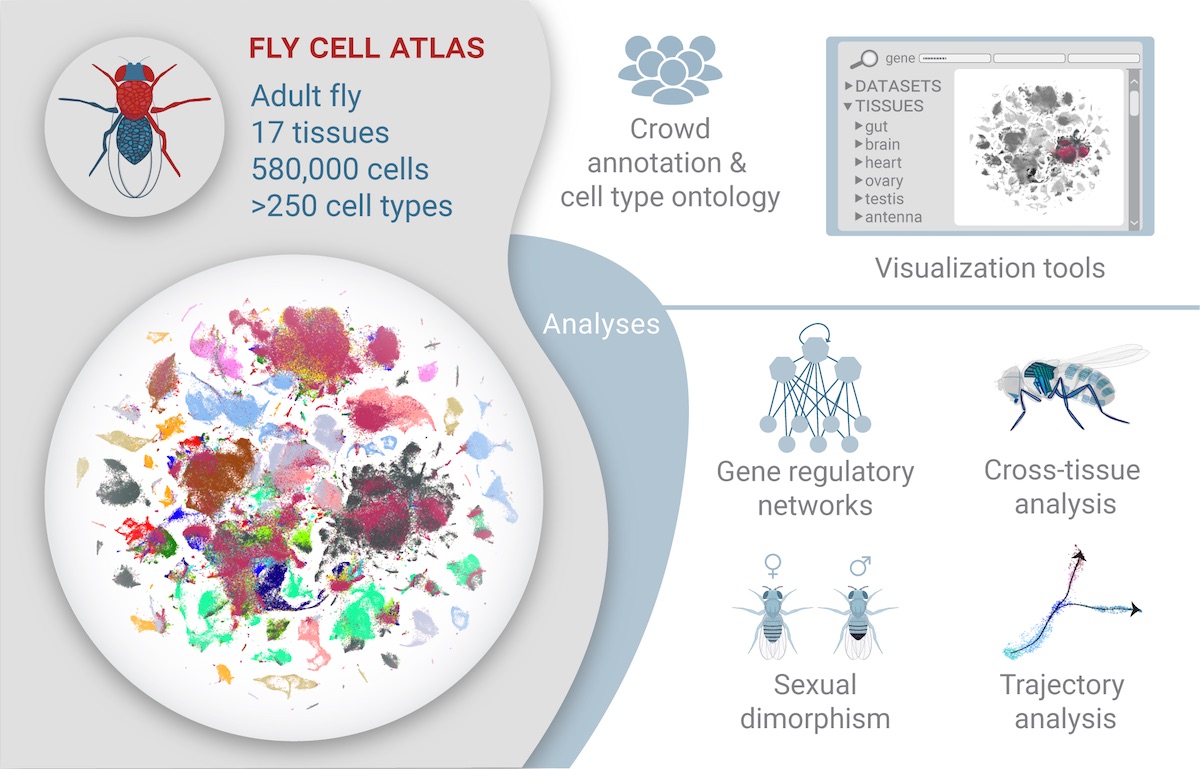
Visual summary of the study published in Science
This ambitious, multi-year project was spearheaded by the labs of Wu Tsai Neuro affiliates Liqun Luo and Stephen Quake at Stanford and the Chan Zuckerberg Biohub (CZ Biohub), along with Stein Aerts at KU Leuven in Belgium, Bart Deplancke at Ecole Polytechnique Fédérale de Lausann (EPFL) in Switzerland, and Norbert Perrimon at Harvard. It was led by Hongjie Li, a former Wu Tsai Neuro Interdisciplinary Postdoctoral Scholar in the Luo and Quake labs at Stanford, who is now an assistant professor at Baylor University, and graduate student Jasper Janssens at KU Leuven.
Li and colleagues at Stanford and the CZ Biohub sequenced 580,000 cells from 15 dissected tissues collected from many laboratories, as well as whole fly heads and bodies. The KU Leuven team organized data analysis pipelines and crowdsourced cell-type annotation across 40 labs, with over 100 experts assigning cell types. Altogether, the authors identified 17 cell categories and 251 specific cell types, many of which were described for the first time. A number of Drosophila tissues—including the heart, leg, proboscis, and wings—had never before been single-cell sequenced prior to this work.
“The Fly Cell Atlas provides the most comprehensive dataset for genome-wide expression in most cell types, which will facilitate the study of many branches of biology using the fruit fly as a model organism,” said Luo, who is Ann and Bill Swindells Professor in the Department of Biology, a member of the Wu Tsai Neurosciences Institute, and a Howard Hughes Medical Institute Investigator.
“I am so pleased that the CZ Biohub was able to contribute to this monumental community resource,” said Quake, who is Lee Otterson Professor of bioengineering and of applied physics and a member of the Wu Tsai Neurosciences Institute at Stanford, and president of the CZ Biohub Network. “We are excited that whole-organism cell atlases are now reaching fruition for a number of important organisms and are being made available to scientists across the globe.”
Luo and Quake are among the leaders of the Wu Tsai Neurosciences Institute Neuro-Omics Initiative, which aims to use 'omics techniques to understand the brain from molecules to brain circuits and across evolutionary time.
Learn more
More fly atlases are in the works, the researchers say: from developing to aging flies.
Importantly, all data and bioinformatics pipelines from this and ongoing studies are publicly available on the FCA website for researchers to explore and use, Luo explained. “Now, instead of carrying out expensive, time-consuming single-cell sequencing, scientists around the world can just look up the relevant information in the FCA database to drive new hypotheses and novel experiments.”

